Reshaping Data
Data reshaping is a crucial skill in ecological research, where we often need to transform our data between different formats to facilitate analysis or visualization. This article will introduce you to the fundamental concepts of data reshaping in R using the tidyverse package, focusing on two main operations: pivot_longer and pivot_wider.
Introduction
Why reshape and what is tidy data?
In ecological research, data often comes in different formats. Sometimes we have wide data, where each row represents a unique observation (like a plot) and columns represent different measurements or time points. Other times, we have long data, where each row represents a single measurement, and we have columns that identify what that measurement belongs to.
Tidy data is a standardized way of organizing data that makes it easier to analyze. In tidy data:
- Each variable forms a column
- Each observation forms a row
Reshaping data is essential because different analyses and visualizations require different data formats. For example, time series analysis often works better with wide data, while many statistical tests and ggplot2 visualizations work better with long data.
Pivot Longer
Pivot longer transforms wide data into long data by collapsing multiple columns into two new columns: one for the variable names and one for their values. This is particularly useful when you have repeated measurements or multiple variables stored in column names.
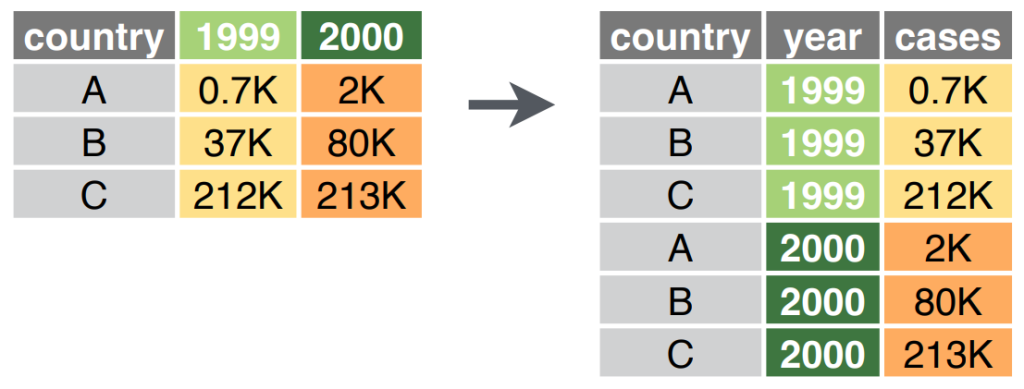
Let’s start with a dataset for this:
df_wide_eco <- tibble(
# Plot information
plot_id = paste0("Plot", str_pad(1:6, 2, pad = "0")),
plot_type = rep(c("Control", "Treatment"), 3),
elevation = round(rnorm(6, mean = 500, sd = 50), 0), # meters
# Quercus rubra measurements for 2020-2022
Q_rubra_height_2020 = round(rnorm(6, mean = 15, sd = 2), 1),
Q_rubra_height_2021 = round(rnorm(6, mean = 16, sd = 2), 1),
Q_rubra_height_2022 = round(rnorm(6, mean = 17, sd = 2), 1),
Q_rubra_dbh_2020 = round(rnorm(6, mean = 30, sd = 5), 1),
Q_rubra_dbh_2021 = round(rnorm(6, mean = 32, sd = 5), 1),
Q_rubra_dbh_2022 = round(rnorm(6, mean = 34, sd = 5), 1),
# Acer saccharum measurements for 2020-2022
A_saccharum_height_2020 = round(rnorm(6, mean = 12, sd = 1.5), 1),
A_saccharum_height_2021 = round(rnorm(6, mean = 13, sd = 1.5), 1),
A_saccharum_height_2022 = round(rnorm(6, mean = 14, sd = 1.5), 1),
A_saccharum_dbh_2020 = round(rnorm(6, mean = 25, sd = 4), 1),
A_saccharum_dbh_2021 = round(rnorm(6, mean = 27, sd = 4), 1),
A_saccharum_dbh_2022 = round(rnorm(6, mean = 29, sd = 4), 1),
# Betula alleghaniensis measurements for 2020-2022
B_alleghaniensis_height_2020 = round(rnorm(6, mean = 18, sd = 2.5), 1),
B_alleghaniensis_height_2021 = round(rnorm(6, mean = 19, sd = 2.5), 1),
B_alleghaniensis_height_2022 = round(rnorm(6, mean = 20, sd = 2.5), 1),
B_alleghaniensis_dbh_2020 = round(rnorm(6, mean = 35, sd = 6), 1),
B_alleghaniensis_dbh_2021 = round(rnorm(6, mean = 37, sd = 6), 1),
B_alleghaniensis_dbh_2022 = round(rnorm(6, mean = 39, sd = 6), 1)
)
df_wide_eco# A tibble: 6 × 21
plot_id plot_type elevation Q_rubra_height_2020 Q_rubra_height_2021 Q_rubra_height_2022 Q_rubra_dbh_2020 Q_rubra_dbh_2021 Q_rubra_dbh_2022 A_saccharum_height_2020 A_saccharum_height_2021 A_saccharum_height_2022 A_saccharum_dbh_2020 A_saccharum_dbh_2021 A_saccharum_dbh_2022 B_alleghaniensis_height_2020 B_alleghaniensis_heigh…¹ B_alleghaniensis_hei…² B_alleghaniensis_dbh…³ B_alleghaniensis_dbh…⁴
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Plot01 Control 490 15.8 15.1 17 32.4 28.8 30.1 11 12.2 13.4 32.1 36.6 30.2 12.4 17.4 23.6 41.7 40.1
2 Plot02 Treatment 533 13.1 17.7 13.4 36.9 21.2 38.2 12 13.3 10 26.1 27 25 21.1 19.4 22.6 32.1 34.7
3 Plot03 Control 514 16.7 17 17.1 32.3 36.4 27.7 13 13.7 13.9 25.5 33.5 29.1 14.9 20.1 21.1 36.4 30.4
4 Plot04 Treatment 551 14.1 14.8 17.4 24.3 27.9 32.2 9.5 13.4 14.6 30.1 21.2 24.7 19.1 21.2 21.8 33.2 44.3
5 Plot05 Control 541 19.8 14 17.3 27.8 29.1 33.6 11.5 14 14.8 22.1 26.2 31.9 19.6 13.9 22.3 40.2 41.4
6 Plot06 Treatment 490 11.7 16.3 14.9 31.7 39.5 28.2 13.1 12.8 13.2 23.2 28.5 33.3 17.5 14.9 13.3 32.9 47.3
# ℹ abbreviated names: ¹B_alleghaniensis_height_2021, ²B_alleghaniensis_height_2022, ³B_alleghaniensis_dbh_2020, ⁴B_alleghaniensis_dbh_2021
# ℹ 1 more variable: B_alleghaniensis_dbh_2022 <dbl>The basic syntax of pivot_longer requires three main arguments:
cols: Specifies which columns to pivot (can use tidyselect helpers likestarts_with())names_to: The name of the new column that will contain the old column namesvalues_to: The name of the new column that will contain the values
Example 1: Basic Pivot Longer
df_long_simple <- df_wide_eco %>%
pivot_longer(
cols = starts_with(c("Q_", "A_", "B_")),
names_to = "target",
values_to = "value"
)
df_long_simple# A tibble: 108 × 5
plot_id plot_type elevation target value
<chr> <chr> <dbl> <chr> <dbl>
1 Plot01 Control 490 Q_rubra_height_2020 15.8
2 Plot01 Control 490 Q_rubra_height_2021 15.1
3 Plot01 Control 490 Q_rubra_height_2022 17
4 Plot01 Control 490 Q_rubra_dbh_2020 32.4
5 Plot01 Control 490 Q_rubra_dbh_2021 28.8
6 Plot01 Control 490 Q_rubra_dbh_2022 30.1
7 Plot01 Control 490 A_saccharum_height_2020 11
8 Plot01 Control 490 A_saccharum_height_2021 12.2
9 Plot01 Control 490 A_saccharum_height_2022 13.4
10 Plot01 Control 490 A_saccharum_dbh_2020 32.1
# ℹ 98 more rowsIn this example, we’ve taken all columns that start with “Q_”, “A_”, or “B_” (our species measurements) and collapsed them into two new columns: “target” (containing the original column names) and “value” (containing the measurements). This gives us a long-format dataset where each row represents a single measurement.
The names_sep argument allows us to split the column names into multiple columns based on a delimiter. This is useful when your column names contain multiple pieces of information separated by a character (like an underscore).
Example 2: Using names_sep to pivot longer & splitting the singular collapsed column into multiple
df_long_split <- df_wide_eco %>%
pivot_longer(
cols = starts_with(c("Q_", "A_", "B_")),
names_sep = "_",
names_to = c("genus_letter", "species_code", "measurement_type", "survey_year"),
values_to = "measurement_value"
)
df_long_split# A tibble: 108 × 8
plot_id plot_type elevation genus_letter species_code measurement_type survey_year measurement_value
<chr> <chr> <dbl> <chr> <chr> <chr> <chr> <dbl>
1 Plot01 Control 490 Q rubra height 2020 15.8
2 Plot01 Control 490 Q rubra height 2021 15.1
3 Plot01 Control 490 Q rubra height 2022 17
4 Plot01 Control 490 Q rubra dbh 2020 32.4
5 Plot01 Control 490 Q rubra dbh 2021 28.8
6 Plot01 Control 490 Q rubra dbh 2022 30.1
7 Plot01 Control 490 A saccharum height 2020 11
8 Plot01 Control 490 A saccharum height 2021 12.2
9 Plot01 Control 490 A saccharum height 2022 13.4
10 Plot01 Control 490 A saccharum dbh 2020 32.1
# ℹ 98 more rowsHere, we’ve used names_sep = "_" to split the column names at each underscore, creating four new columns: “genus_letter” (Q, A, B), “species_code” (rubra, saccharum, alleghaniensis), “measurement_type” (height, dbh), and “survey_year” (2020, 2021, 2022). This gives us a much more structured dataset where each piece of information from the column names is in its own column.
The names_pattern argument provides even more control over how column names are split. It uses regular expressions (regex) to extract specific parts of the column names. This is particularly useful when your column names follow a complex pattern or when you need more precise control over the splitting.
Example 3: Using names_pattern to perform a more fine-tuned splitting on the singular collapsed column
df_long_pattern <- df_wide_eco %>%
pivot_longer(
cols = starts_with(c("Q_", "A_", "B_")),
names_to = c("sample_name", "measurement_type", "survey_year"),
names_pattern = "([A-Z]_[a-z]+)_([a-z]+)_(\\d{4})",
values_to = "measurement_value"
)
df_long_pattern# A tibble: 108 × 7
plot_id plot_type elevation sample_name measurement_type survey_year measurement_value
<chr> <chr> <dbl> <chr> <chr> <chr> <dbl>
1 Plot01 Control 490 Q_rubra height 2020 15.8
2 Plot01 Control 490 Q_rubra height 2021 15.1
3 Plot01 Control 490 Q_rubra height 2022 17
4 Plot01 Control 490 Q_rubra dbh 2020 32.4
5 Plot01 Control 490 Q_rubra dbh 2021 28.8
6 Plot01 Control 490 Q_rubra dbh 2022 30.1
7 Plot01 Control 490 A_saccharum height 2020 11
8 Plot01 Control 490 A_saccharum height 2021 12.2
9 Plot01 Control 490 A_saccharum height 2022 13.4
10 Plot01 Control 490 A_saccharum dbh 2020 32.1
# ℹ 98 more rowsIn this example, we’ve used a regular expression pattern to extract three groups from the column names: the species name (e.g., “Q_rubra”), the measurement type (e.g., “height”), and the year (e.g., “2020”). The pattern ([A-Z]_[a-z]+)_([a-z]+)_(\\d{4}) breaks down as:
([A-Z]_[a-z]+)– First group: A capital letter followed by an underscore and lowercase letters (species name)_([a-z]+)_– Second group: An underscore, lowercase letters, and another underscore (measurement type)(\\d{4})– Third group: Four digits (year)
This gives us more control over how the column names are split compared to names_sep, allowing us to keep the species name as a single column rather than splitting it into genus and species.
It is understandable that regular expressions can feel like an alien language at this point for you. Fortunately, if you have the time, there are fantastic resources online such as this start-to-finish tutorial that will get you familiar with regular expressions in no time!
Pivot Wider
Pivot wider is the opposite of pivot longer – it transforms long data into wide data by spreading a column of variable names into multiple columns. This is useful when you want to create a summary table or when you need to prepare data for certain types of analysis.
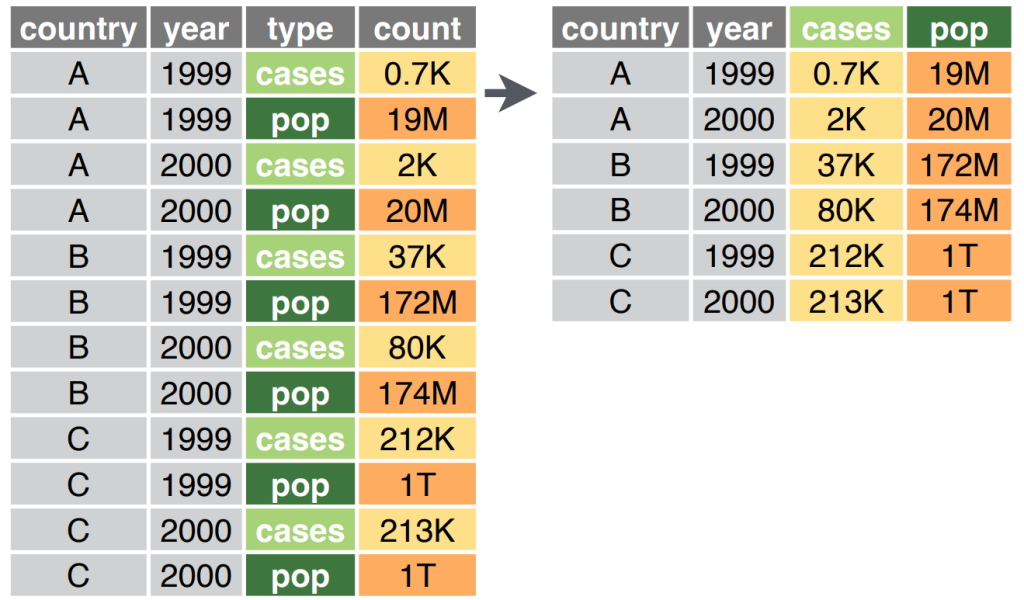
We’ll make a dataset for this:
df_long_eco <- tibble(
plot_id = rep(paste0("Plot", str_pad(1:4, 2, pad = "0")), each = 9), # 4 plots × 3 species × 3 years
species = rep(rep(c("Quercus rubra", "Acer saccharum", "Betula alleghaniensis"), each = 3), 4),
year = rep(2020:2022, 12), # 12 combinations of plot × species
height = round(rnorm(36, mean = 15, sd = 2), 1), # Height in meters
dbh = round(rnorm(36, mean = 30, sd = 5), 1), # Diameter at breast height in cm
leaf_area = round(rnorm(36, mean = 100, sd = 15), 1) # Leaf area in cm²
)
df_long_eco# A tibble: 36 × 6
plot_id species year height dbh leaf_area
<chr> <chr> <int> <dbl> <dbl> <dbl>
1 Plot01 Quercus rubra 2020 14.3 28.9 92.2
2 Plot01 Quercus rubra 2021 15.2 30.8 110.
3 Plot01 Quercus rubra 2022 18.2 35.8 99.1
4 Plot01 Acer saccharum 2020 14.8 35.3 110.
5 Plot01 Acer saccharum 2021 17.2 35.7 120
6 Plot01 Acer saccharum 2022 16.3 27.1 100.
7 Plot01 Betula alleghaniensis 2020 14.8 40 115.
8 Plot01 Betula alleghaniensis 2021 11.9 30.3 82.2
9 Plot01 Betula alleghaniensis 2022 14 39.3 89.2
10 Plot02 Quercus rubra 2020 14 23.2 123.
# ℹ 26 more rows# A tibble: 12 × 5
plot_id species height_2020 height_2021 height_2022
<chr> <chr> <dbl> <dbl> <dbl>
1 Plot01 Quercus rubra 14.3 15.2 18.2
2 Plot01 Acer saccharum 14.8 17.2 16.3
3 Plot01 Betula alleghaniensis 14.8 11.9 14
4 Plot02 Quercus rubra 14 15.1 17.6
5 Plot02 Acer saccharum 19.6 18.1 14.7
6 Plot02 Betula alleghaniensis 11.5 14.2 15.2
7 Plot03 Quercus rubra 16.7 16.9 16.4
8 Plot03 Acer saccharum 12.2 16.7 14.1
9 Plot03 Betula alleghaniensis 15.3 15.1 15.9
10 Plot04 Quercus rubra 15 11.7 16.5
11 Plot04 Acer saccharum 15.8 14.5 15.2
12 Plot04 Betula alleghaniensis 15.3 15.4 18.3The pivot_wider function requires at least two main arguments:
names_from: The column whose values will become the new column namesvalues_from: The column whose values will fill the new columns
Example 1: Basic pivot_wider.
df_wide_basic <- df_long_eco %>%
select(plot_id, species, year, height) %>%
pivot_wider(
names_from = year,
values_from = height,
names_prefix = "height_"
)
df_wide_basicIn this example, we’ve taken the “year” column and used its values (2020, 2021, 2022) to create new column names, prefixed with “height_”. The values from the “height” column are then placed in these new columns. This gives us a wide-format dataset where each row represents a unique combination of plot and species, and the columns represent height measurements for each year.
The names_glue argument provides a template for creating the new column names. It uses a special syntax where {.value} is replaced with the name of the value column, and any other column names in curly braces are replaced with their values. This is particularly useful when you have multiple value columns and want to create more descriptive column names.
Example 2: Using names_glue.
df_wide_glue <- df_long_eco %>%
pivot_wider(
id_cols = c(plot_id, species),
names_from = year,
values_from = c(height, dbh, leaf_area),
names_glue = "{.value}_{year}"
)
df_wide_glue# A tibble: 12 × 11
plot_id species height_2020 height_2021 height_2022 dbh_2020 dbh_2021 dbh_2022 leaf_area_2020 leaf_area_2021 leaf_area_2022
<chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
1 Plot01 Quercus rubra 14.3 15.2 18.2 28.9 30.8 35.8 92.2 110. 99.1
2 Plot01 Acer saccharum 14.8 17.2 16.3 35.3 35.7 27.1 110. 120 100.
3 Plot01 Betula alleghaniensis 14.8 11.9 14 40 30.3 39.3 115. 82.2 89.2
4 Plot02 Quercus rubra 14 15.1 17.6 23.2 30.1 36.2 123. 106. 69.2
5 Plot02 Acer saccharum 19.6 18.1 14.7 26.4 26.2 25.3 79.5 97 113
6 Plot02 Betula alleghaniensis 11.5 14.2 15.2 24.7 27.8 31.7 98.5 109. 114.
7 Plot03 Quercus rubra 16.7 16.9 16.4 19.9 31.1 36.2 125. 101. 99.2
8 Plot03 Acer saccharum 12.2 16.7 14.1 40.2 36.5 33.8 73.7 102. 91.4
9 Plot03 Betula alleghaniensis 15.3 15.1 15.9 21.4 27 28.2 85.4 97.3 115.
10 Plot04 Quercus rubra 15 11.7 16.5 33.5 29.5 23.7 70.1 93.6 102.
11 Plot04 Acer saccharum 15.8 14.5 15.2 38.4 34.6 31.2 86.6 105 106.
12 Plot04 Betula alleghaniensis 15.3 15.4 18.3 36.1 23.3 33.3 99.5 63 139. In this example, we’ve used names_glue = "{.value}_{year}" to create column names that combine the value column name (height, dbh, or leaf_area) with the year. This gives us a wide-format dataset with columns like “height_2020”, “dbh_2020”, “leaf_area_2020”, etc. The id_cols argument specifies which columns should remain as identifiers (plot_id and species in this case).
Summary
Data reshaping is a fundamental skill in ecological data analysis. By mastering pivot_longer and pivot_wider, you can:
- Transform data between wide and long formats to suit different analytical needs
- Extract information embedded in column names into separate columns
- Create summary tables with measurements across time points or treatments
- Prepare data for visualization with ggplot2 or statistical analysis
Remember that tidy data principles guide these operations: each variable should form a column, each observation should form a row, and each type of observational unit should form a table. By reshaping your data appropriately, you can make your analyses more straightforward and your code more readable.